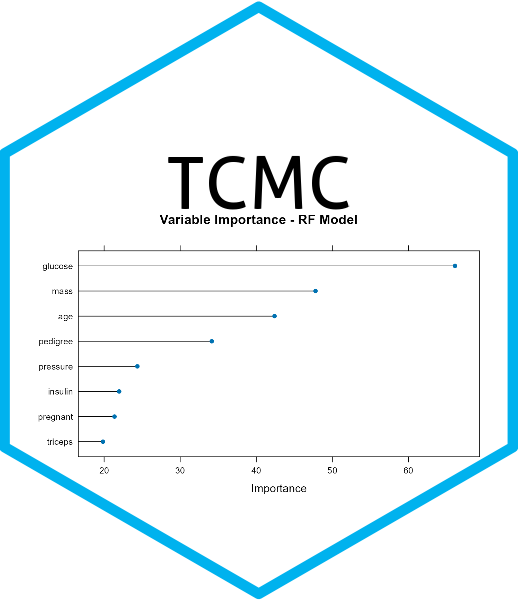
This function compares 13 different classification models using the caret package.
Format
The most important input arguments needed are data and
target_var:
- data
contains 768 rows (observations) and 9 columns (features).
- target_var
column containing a binary vector where 1 indicates diabetes patients and 0 for otherwise.
Arguments
- data
A data frame containing the dataset to be used for modeling.
- target_var
The name of the target variable in the dataset.
- train_prop
The proportion of data to be used for training (default is 0.8).
- seed
The random seed for reproducibility (default is 3456).
- for_utest
only for unit test when is TRUE (FALSE by default).
Details
This data set utilized in the example is originally from the National Institute of Diabetes and Digestive and Kidney Diseases.