Run Genomic Case Study: DNA Sequence Diversity
Source:R/biological_example.R
run_genomic_case_study.RdCase study analyzing genetic diversity using decoupling of Hamming distances
Examples
# Run case study with default parameters
result <- run_genomic_case_study()
#>
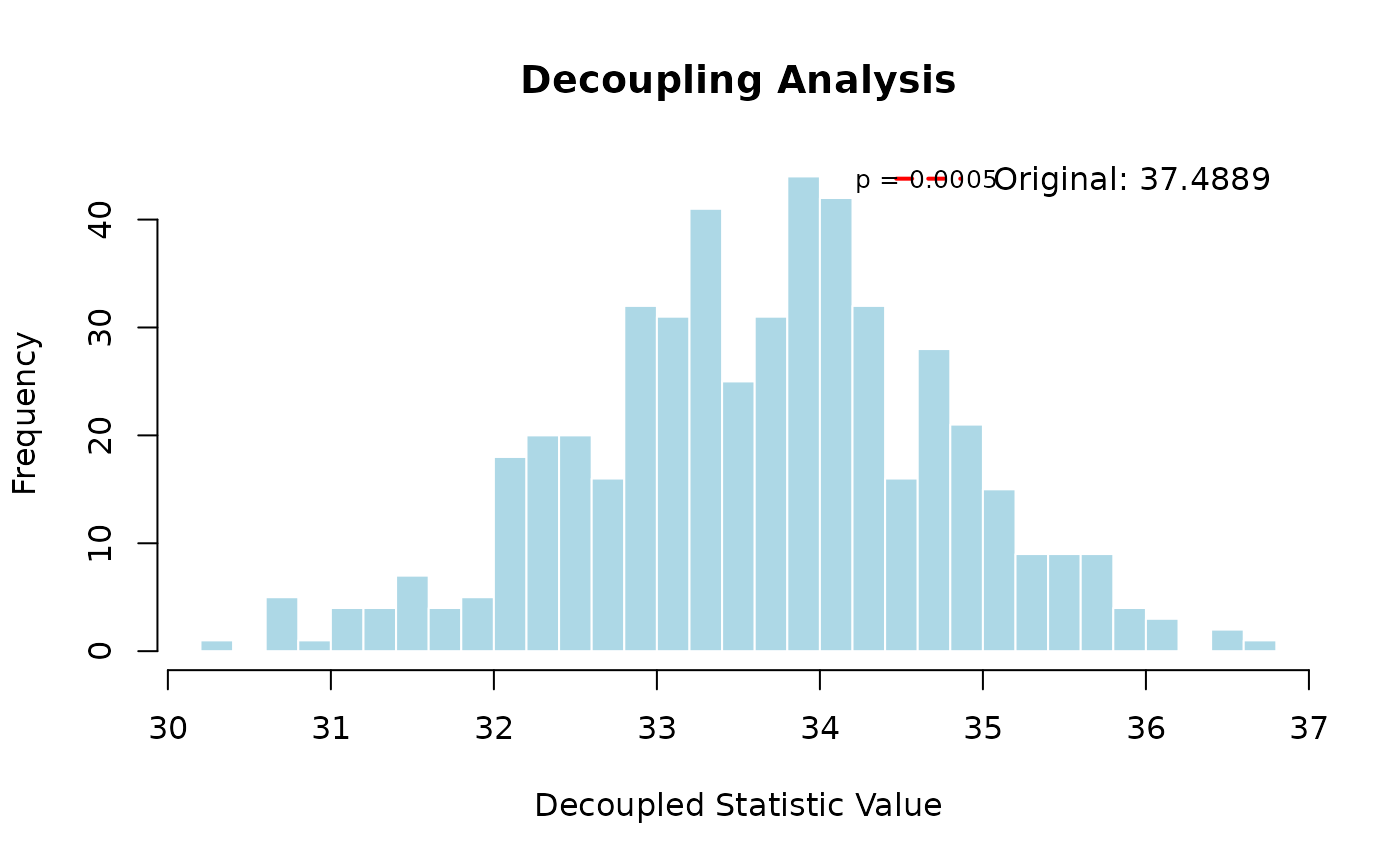
#> === Biological Interpretation ===
#> Original mean Hamming distance: 37.4889
#> Expected distance under independence: 33.6505
#> Observed distance is 3.46 standard deviations from independence expectation
#> Significant evidence of dependence between sequences (p < 0.05)
#> This suggests shared evolutionary history or functional constraints
# Plot results
plot(result)