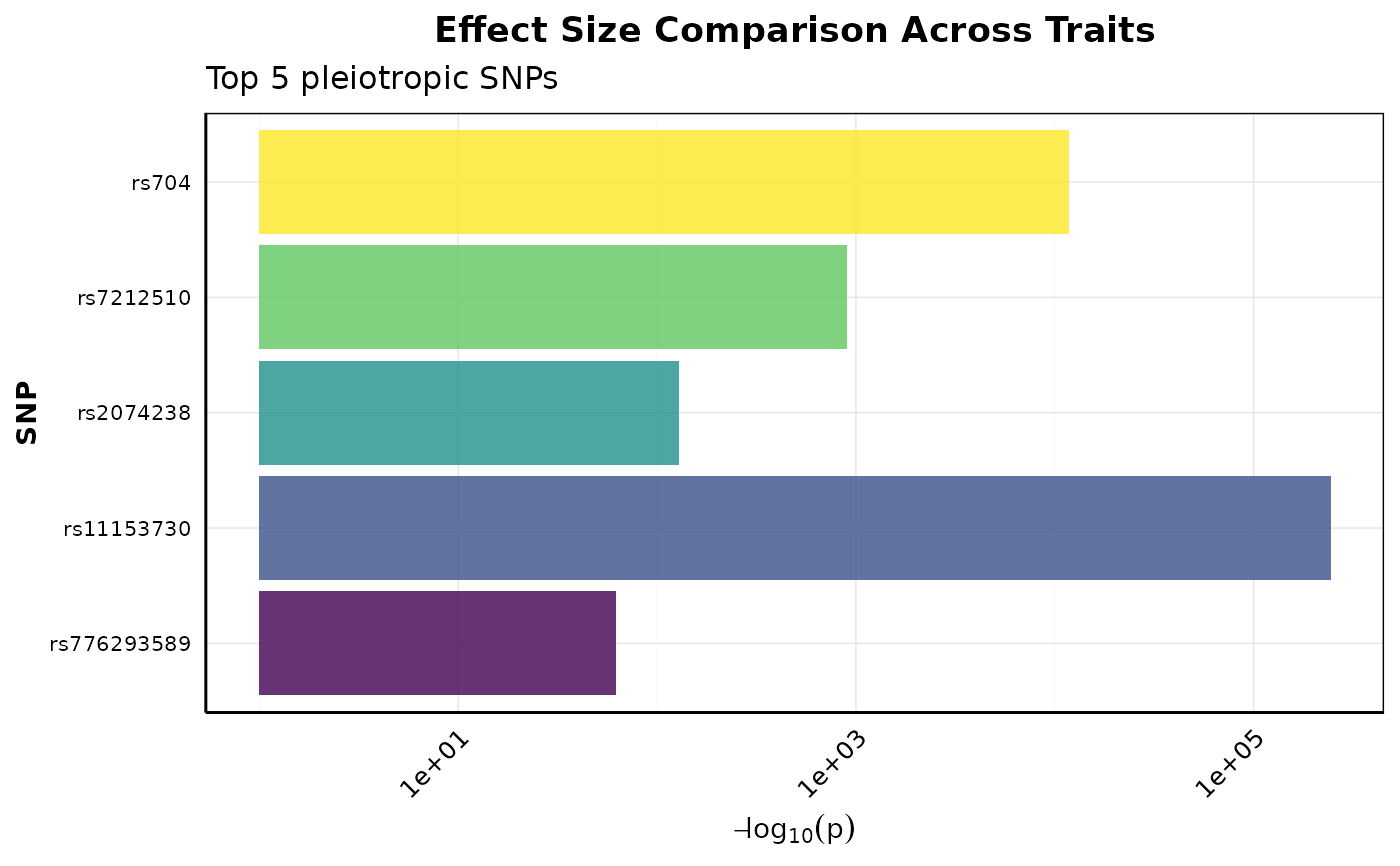
Creates a comparison plot showing effect sizes across traits for pleiotropic SNPs. Helps identify which SNPs have stronger effects on specific traits.
Usage
plot_effect_size_comparison(
pleio_data,
top_n_snps = 15,
effect_col = "PVALUE_MLOG",
use_log_scale = TRUE,
show_error_bars = FALSE,
error_col = NULL,
color_by = "snp",
flip_coords = TRUE
)Arguments
- pleio_data
A data.frame containing pleiotropy analysis results
- top_n_snps
Integer. Number of top pleiotropic SNPs to include (default: 15)
- effect_col
Character. Column to use for effect size (default: "PVALUE_MLOG")
- use_log_scale
Logical. Use log scale for effect sizes (default: TRUE)
- show_error_bars
Logical. Show error bars (default: FALSE)
- error_col
Character. Column for standard errors (default: NULL)
- color_by
Character. Color by: "snp", "trait", "chromosome" (default: "snp")
- flip_coords
Logical. Flip x and y coordinates (default: TRUE for horizontal bars)
Examples
data(gwas_subset)
pleio_results <- detect_pleiotropy(gwas_subset)
if (nrow(pleio_results) > 0) {
p <- plot_effect_size_comparison(pleio_results, top_n_snps = 5)
print(p)
}