This vignette demonstrates the advanced visualization capabilities of pleior for analyzing and presenting pleiotropic genetic associations. These publication-ready visualizations are designed for scientific journals and provide explainable insights into shared genetic architecture.
Setup
Load the package and example data:
library(pleior)
library(ggplot2)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
data(gwas_subset)Preprocess and detect pleiotropic SNPs:
gwas_clean <- preprocess_gwas(gwas_subset, pvalue_threshold = 5e-8)
pleio_results <- detect_pleiotropy(
gwas_clean |>
dplyr::filter(stringr::str_detect(MAPPED_TRAIT, 'Alzheimer'))
)
pleio_results$MAPPED_TRAIT <- substr(pleio_results$MAPPED_TRAIT, 1, 20)
head(pleio_results)
#> # A tibble: 6 × 7
#> SNPS N_TRAITS TRAITS MAPPED_TRAIT PVALUE_MLOG CHR_ID CHR_POS
#> <chr> <int> <chr> <chr> <dbl> <chr> <chr>
#> 1 rs10119 4 Alzheimer disease;… family hist… 307 19 449034…
#> 2 rs10119 4 Alzheimer disease;… Alzheimer d… 161. 19 449034…
#> 3 rs10119 4 Alzheimer disease;… Alzheimer d… 130. 19 449034…
#> 4 rs10119 4 Alzheimer disease;… late-onset … 56.7 19 449034…
#> 5 rs1038025 2 Alzheimer disease;… Alzheimer d… 83.5 19 449017…
#> 6 rs1038025 2 Alzheimer disease;… Alzheimer d… 39 19 449017…Publication-Ready Themes
pleior includes publication-ready themes optimized for different journals:
p <- ggplot(mtcars, aes(x = wt, y = mpg)) +
geom_point(aes(color = factor(cyl))) +
labs(title = "Example Plot with Publication Theme")
# Default publication theme
p1 <- p + theme_pleiotropy_publication()
# Nature journal style
p2 <- p + theme_pleiotropy_publication(journal_style = "nature")
# Science journal style
p3 <- p + theme_pleiotropy_publication(journal_style = "science")
# Colorblind-friendly palettes
colors <- get_pleiotropy_colors("okabe_ito", n = 5)
print(colors)
#> [1] "#E69F00" "#3CAF66" "#6A6859" "#321E29" "#F0F0F0"Network Graph Visualization
Visualize connections between pleiotropic SNPs and their associated traits:
if (nrow(pleio_results) > 0) {
p_network <- plot_pleiotropy_network(
pleio_results,
top_n_snps = min(10, nrow(pleio_results)),
node_size_snp = 6,
node_size_trait = 10,
layout = "fr",
show_labels = TRUE,
color_palette = "okabe_ito"
) + theme_pleiotropy_publication()
print(p_network)
}
#> Warning: Unknown or uninitialised column: `SNP`.
#> Unknown or uninitialised column: `SNP`.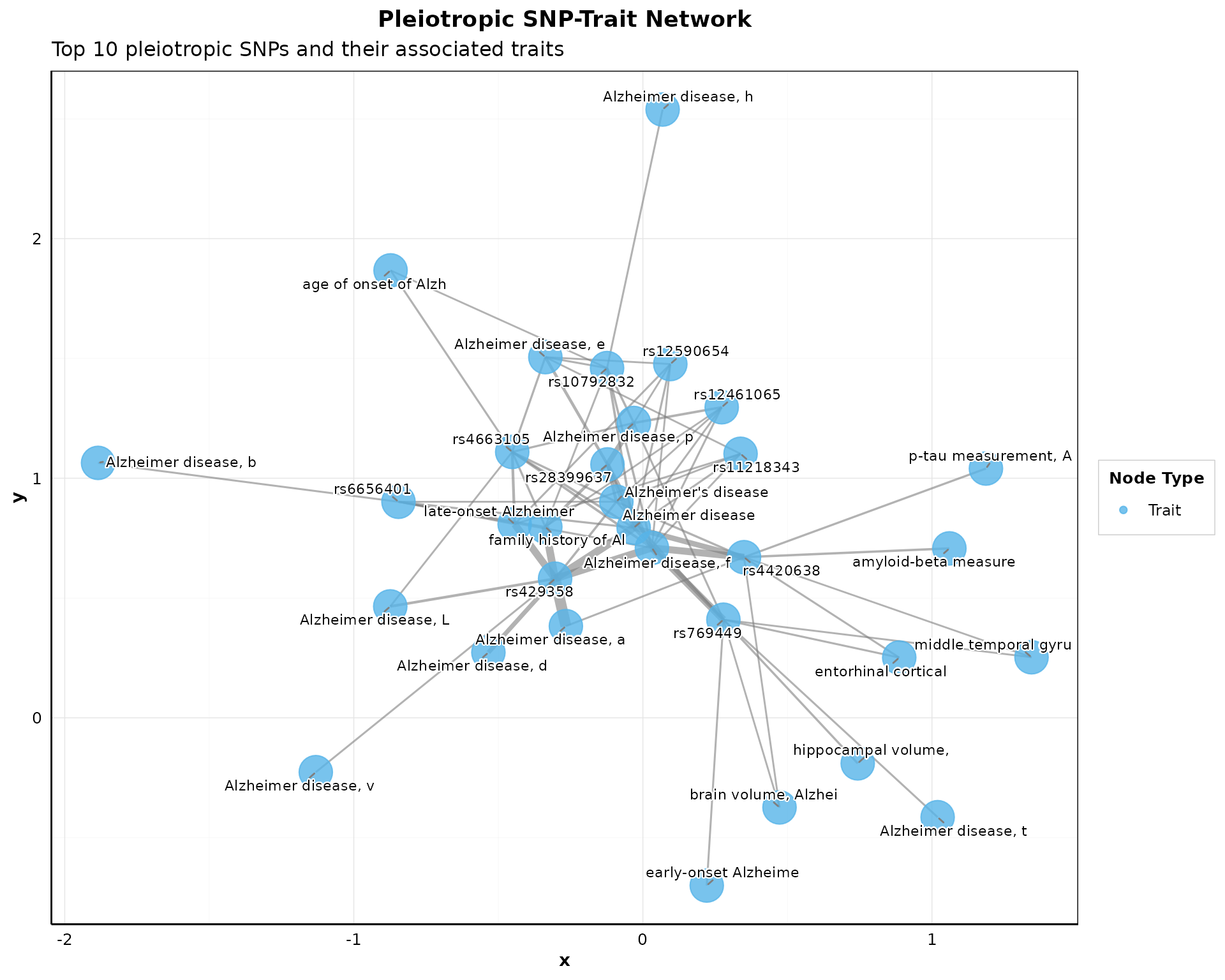
Heatmap Visualization
Create a heatmap showing SNP-trait associations:
if (nrow(pleio_results) > 0) {
p_heatmap <- plot_pleiotropy_heatmap(
pleio_results,
top_n_snps = min(5, nrow(pleio_results)),
top_n_traits = 4,
value_col = "PVALUE_MLOG",
show_dendrograms = FALSE,
color_palette = "bluered",
show_values = TRUE
)
print(p_heatmap)
}
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> ℹ The deprecated feature was likely used in the pleior package.
#> Please report the issue at <https://github.com/danymukesha/pleior/issues>.
#> This warning is displayed once per session.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.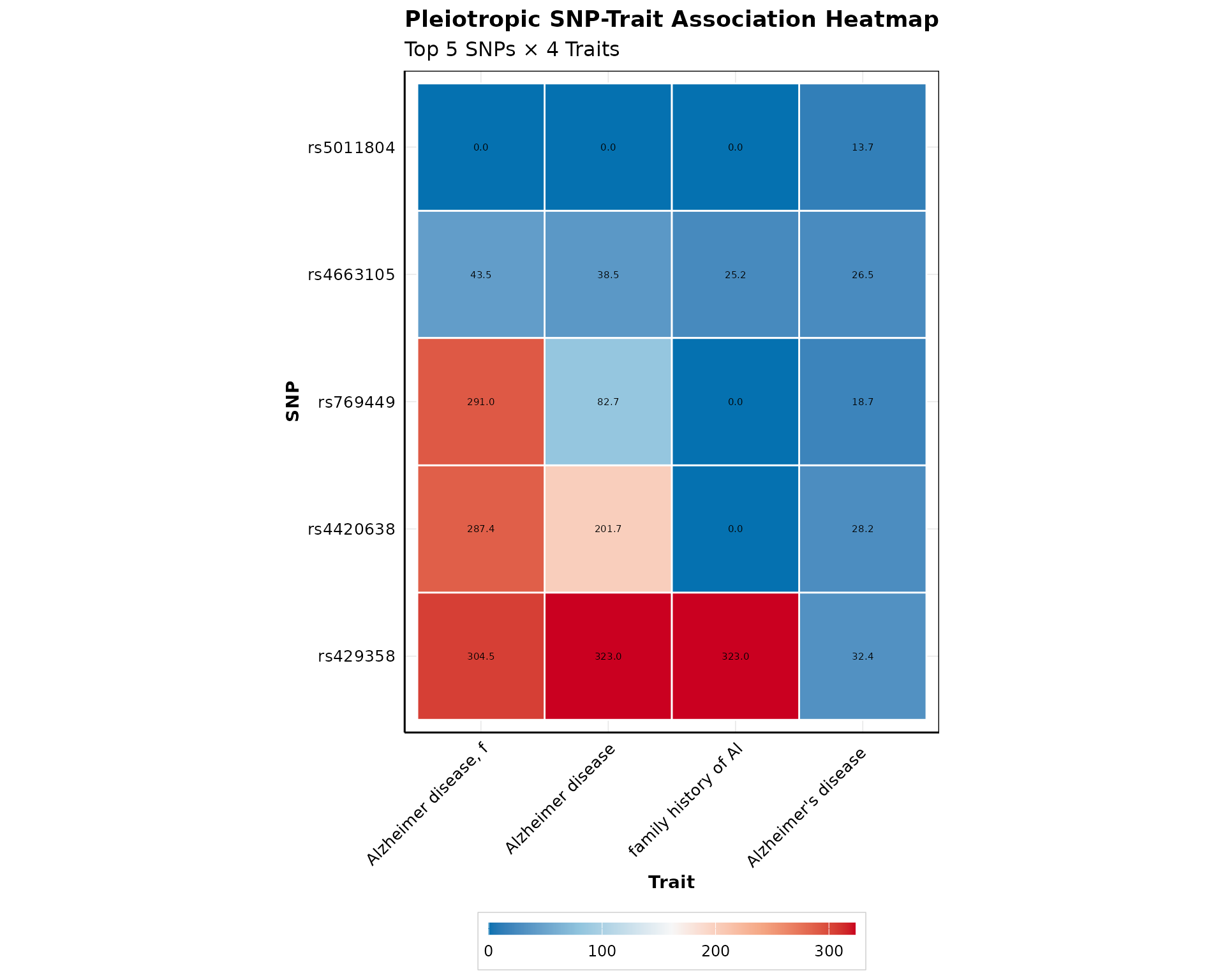
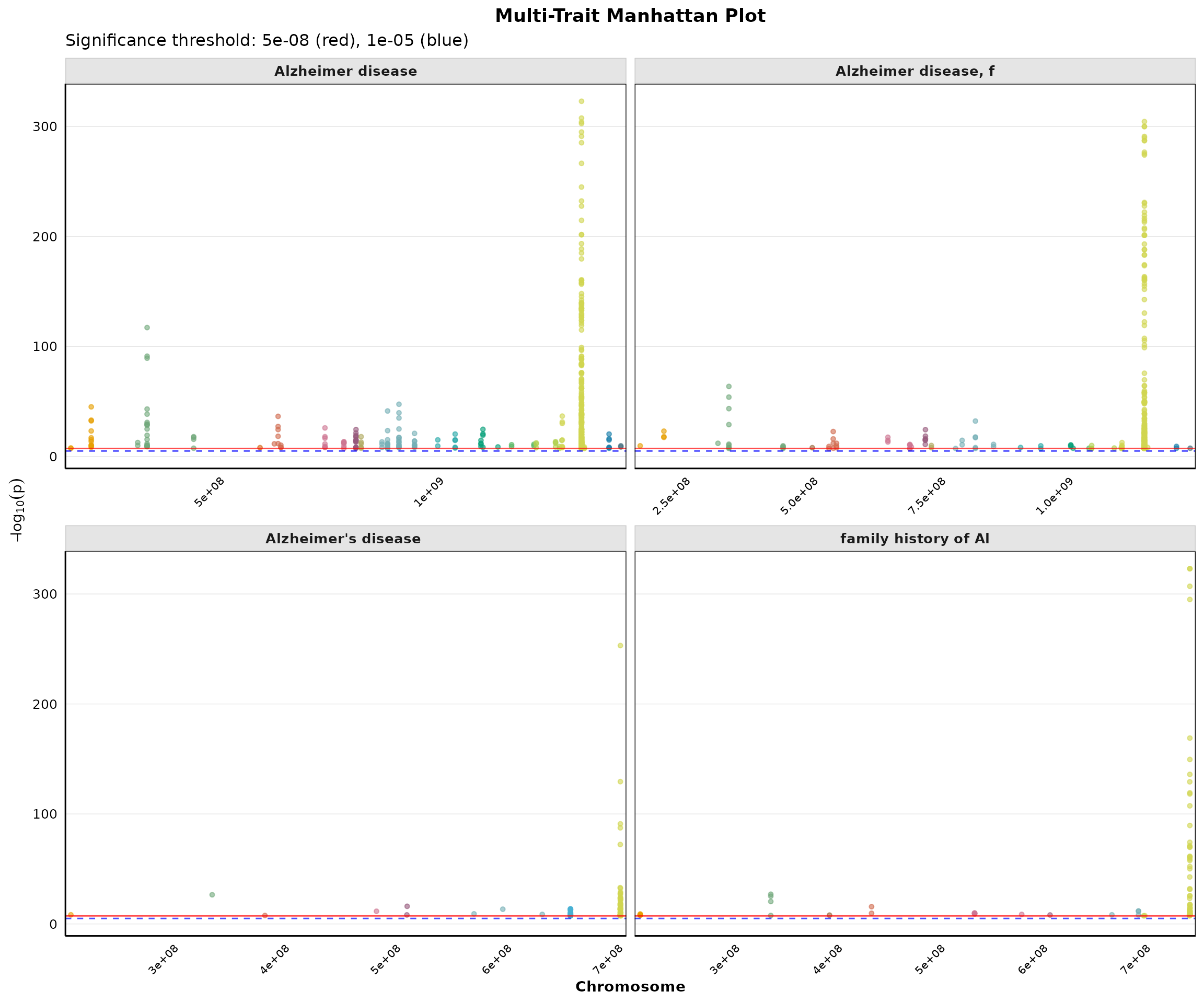
Multi-Trait Manhattan Plot
Compare genome-wide associations across multiple traits:
if (nrow(pleio_results) > 0) {
p_multi <- plot_multi_trait_manhattan(
pleio_results,
max_traits = min(4, length(unique(pleio_results$MAPPED_TRAIT))),
significance_line = 5e-8,
suggestive_line = 1e-5,
point_size = 1.2,
alpha = 0.6,
ncol = 2
)
print(p_multi)
}
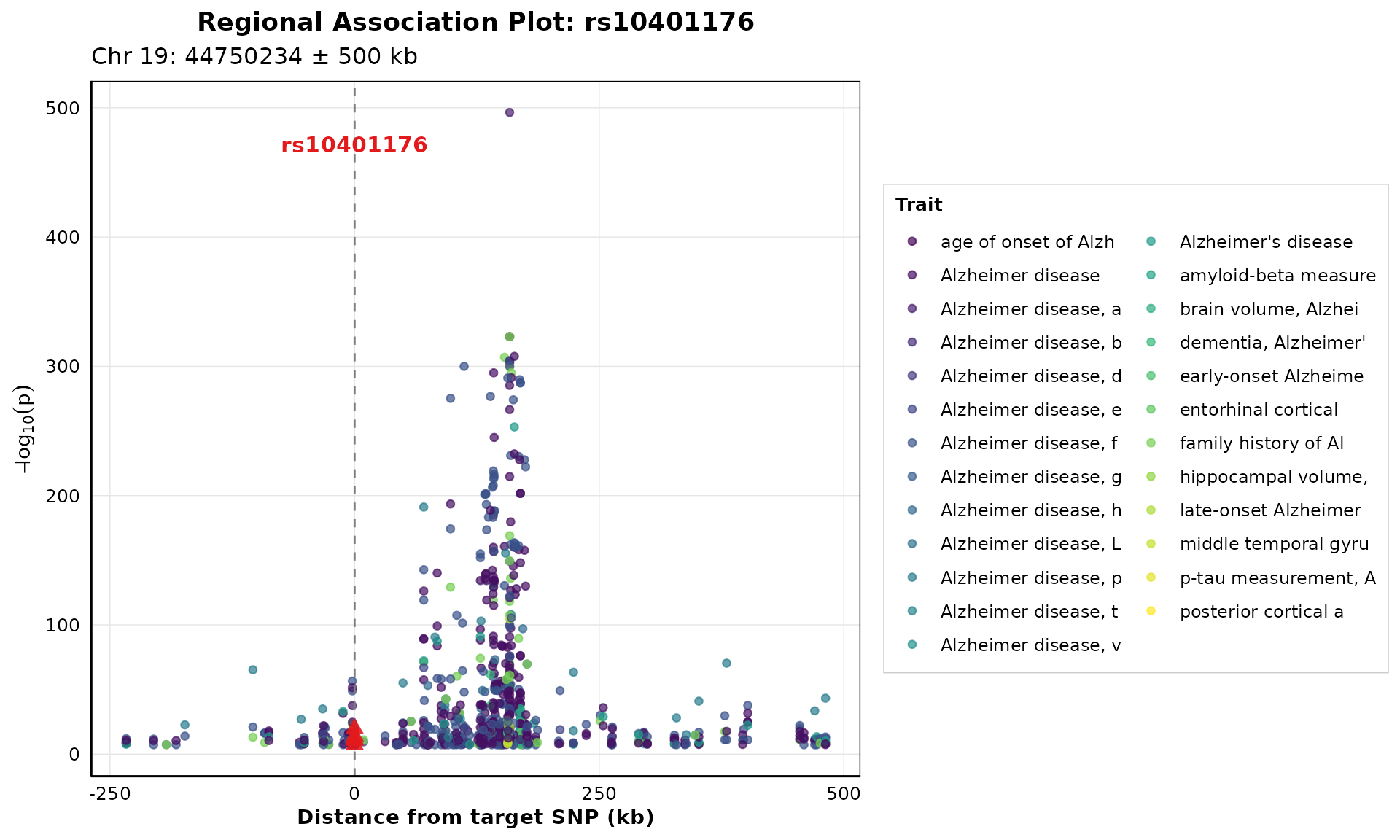
Regional Association Plot
Zoom in on specific genomic regions around pleiotropic SNPs:
if (nrow(pleio_results) > 0 && "rs10401176" %in% pleio_results$SNPS) {
p_regional <- plot_regional_association(
pleio_results,
target_snp = "rs10401176",
window_size = 500000,
highlight_color = "#E31A1C"
)
print(p_regional)
}
Trait Co-occurrence Network
Visualize how traits share pleiotropic SNPs:
if (nrow(pleio_results) > 0) {
p_cooccur <- plot_trait_cooccurrence_network(
pleio_results,
min_shared_snps = 1,
top_n_traits = min(5, length(unique(pleio_results$MAPPED_TRAIT))),
node_size = 8,
layout = "kk",
show_edge_labels = TRUE
# color_by = "n_snps"
)
print(p_cooccur)
}
#> Warning in plot_trait_cooccurrence_network(pleio_results, min_shared_snps = 1,
#> : No trait pairs meet minimum shared SNP criteria
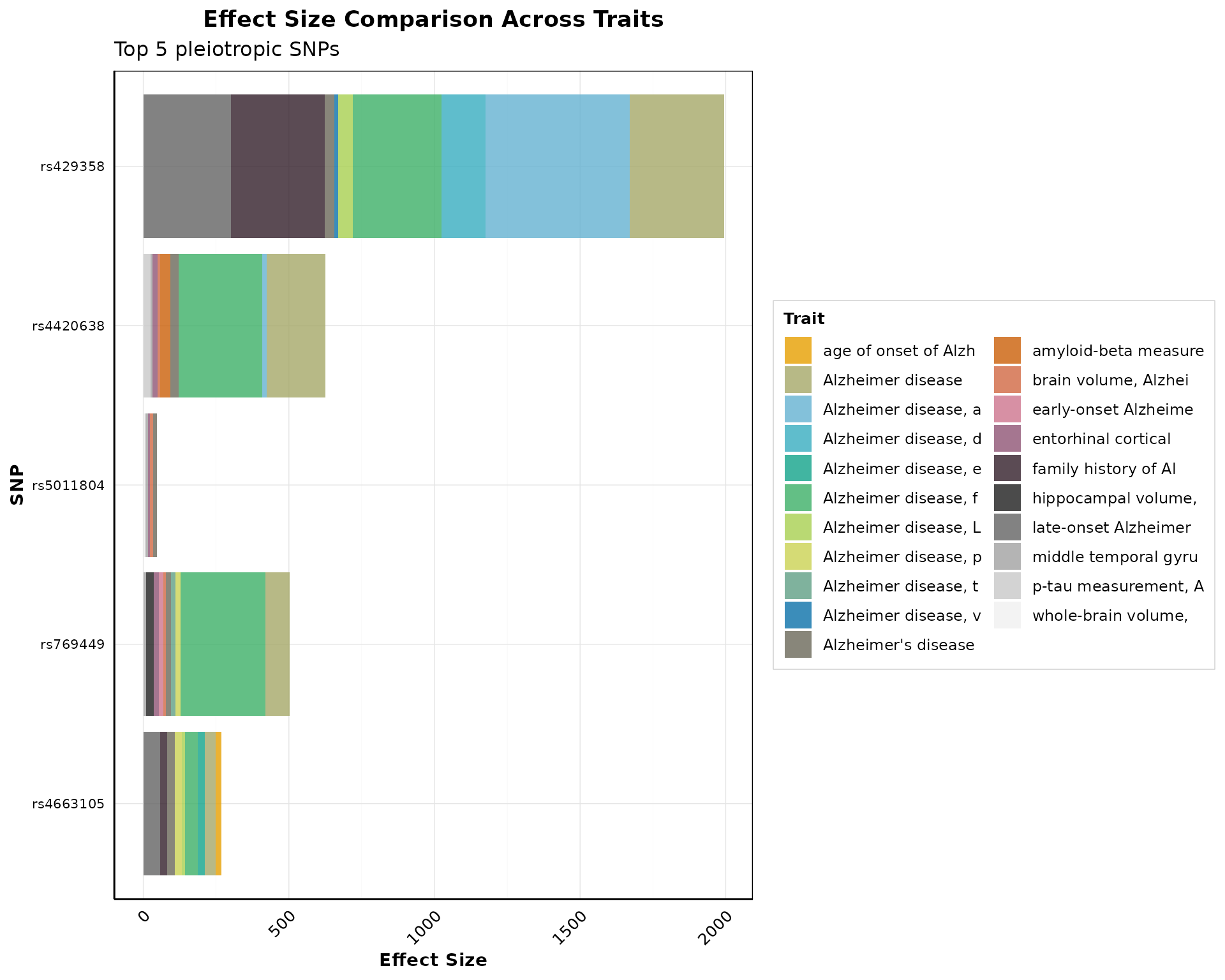
Effect Size Comparison
Compare effect sizes across traits:
if (nrow(pleio_results) > 0) {
p_effect <- plot_effect_size_comparison(
pleio_results,
top_n_snps = min(5, nrow(pleio_results)),
effect_col = "PVALUE_MLOG",
use_log_scale = FALSE,
color_by = "trait",
flip_coords = TRUE
)
print(p_effect)
}
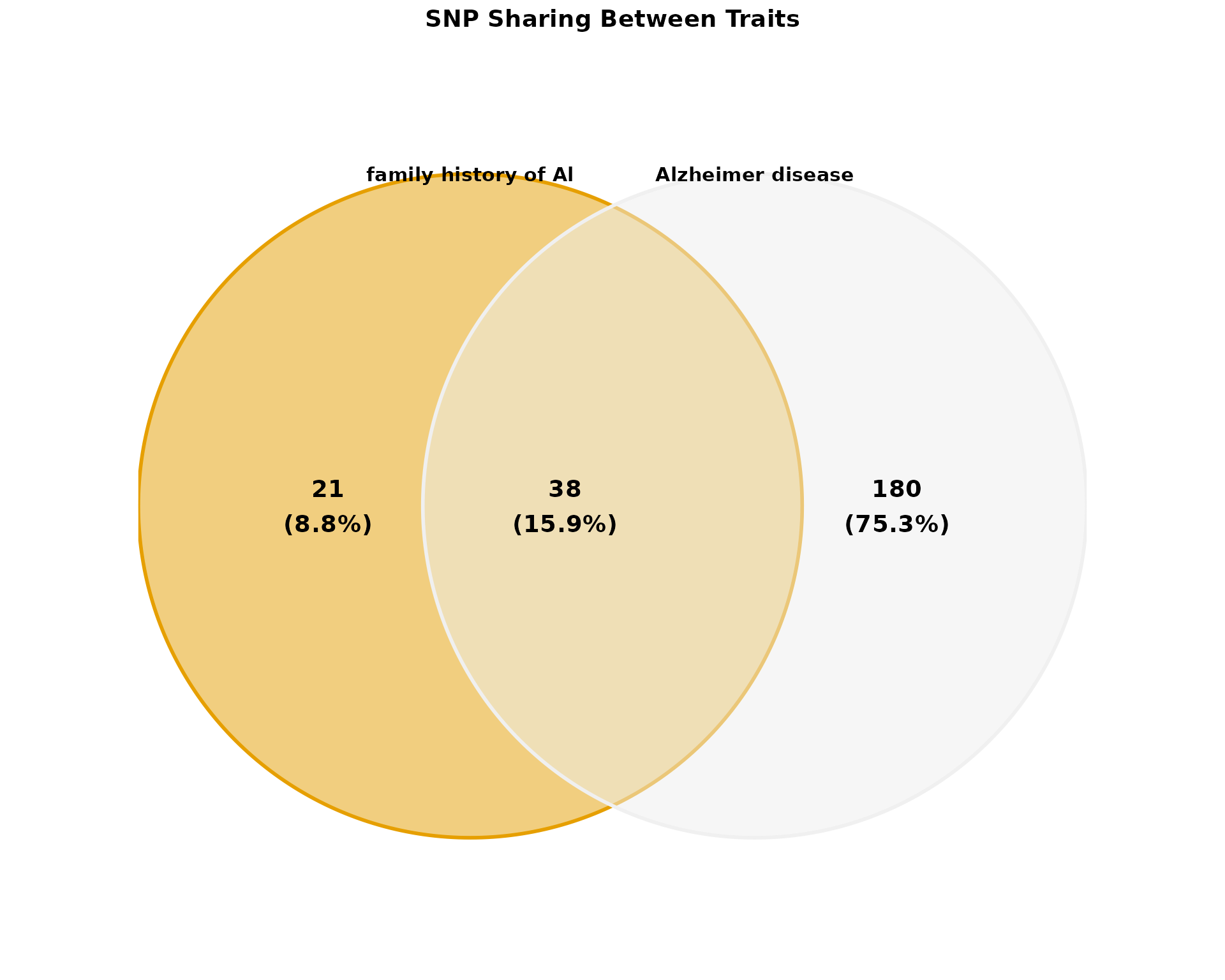
Venn Diagram
Visualize SNP sharing between traits:
if (nrow(pleio_results) > 0) {
traits <- unique(pleio_results$MAPPED_TRAIT)[1:min(3, length(unique(pleio_results$MAPPED_TRAIT)))]
if (length(traits) >= 2) {
p_venn <- plot_venn_diagram(
pleio_results,
traits = traits[1:min(2, length(traits))],
title = "SNP Sharing Between Traits",
show_counts = TRUE,
show_percentages = TRUE,
color_palette = "okabe_ito",
alpha = 0.5
)
print(p_venn)
}
}
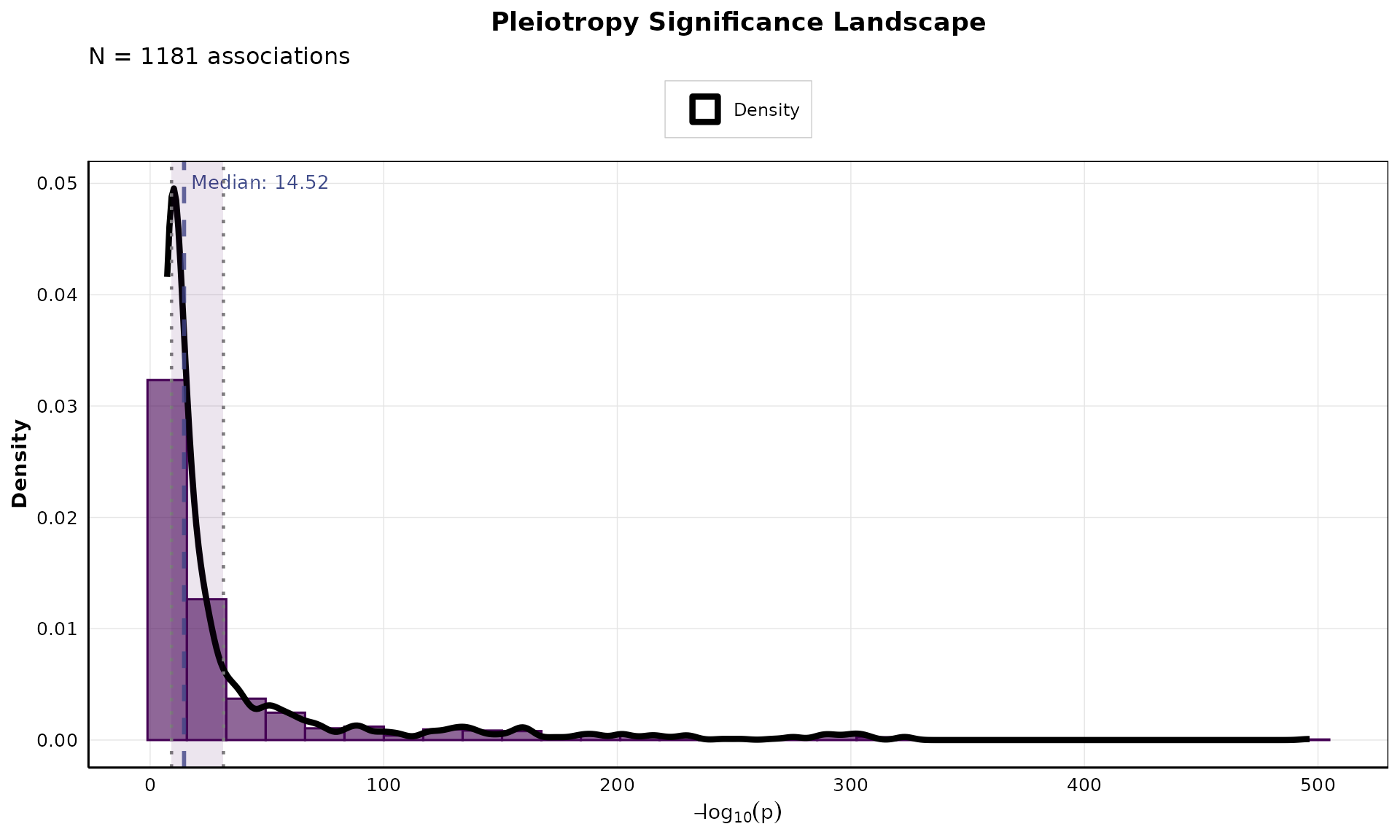
Pleiotropy Significance Landscape
Analyze the distribution of significance values:
if (nrow(pleio_results) > 0) {
p_landscape <- plot_pleiotropy_landscape(
pleio_results,
n_bins = 30,
show_median = TRUE,
show_mean = FALSE,
show_quantiles = TRUE,
color_palette = "viridis"
)
print(p_landscape)
}
Combining Multiple Plots
Create multi-panel figures for publications:
if (nrow(pleio_results) > 0) {
p1 <- plot_pleiotropy_manhattan(
pleio_results,
title = "Manhattan Plot",
) + theme_pleiotropy_publication()
p2 <- plot_pleiotropy_heatmap(
pleio_results,
top_n_snps = min(5, nrow(pleio_results)),
top_n_traits = 4
)
combined <- combine_publication_plots(
p1, p2,
ncol = 2,
labels = c("A", "B")
)
print(combined)
}Saving Publication-Ready Figures
Export figures in high-resolution formats:
# Save as PDF (vector format for journals)
save_publication_plot(
p_heatmap,
"figure1_heatmap.pdf",
width = 8,
height = 6
)
# Save as PNG (high-resolution raster)
save_publication_plot(
p_network,
"figure2_network.png",
width = 10,
height = 8,
dpi = 300
)
# Save as TIFF (very high quality)
save_publication_plot(
p_multi,
"figure3_manhattan.tiff",
width = 12,
height = 10,
dpi = 600
)Customization Options
All visualization functions offer extensive customization:
-
Color palettes: Use
get_pleiotropy_colors()with options like “okabe_ito”, “viridis”, “plasma”, “cividis” -
Journal styles: Apply
theme_pleiotropy_publication()with “nature”, “science”, “pnas”, or “default” - Layout options: Network layouts include “fr”, “kk”, “dh”, “grid”, “circle”
- Highlighting: Emphasize specific SNPs or traits
- Labels and annotations: Add custom labels, titles, and annotations
Best Practices for Publications
- Use vector formats (PDF, SVG) when possible for journals
- Set appropriate DPI (300-600 for raster formats)
- Use colorblind-friendly palettes (e.g., “okabe_ito”)
- Maintain consistent styling across all figures
- Include clear legends and annotations
- Optimize figure dimensions for journal specifications
- Use high-contrast colors for accessibility
For more details, see the function documentation:
