Network Graph Visualization for Pleiotropic SNPs
Source:R/plot_network.R
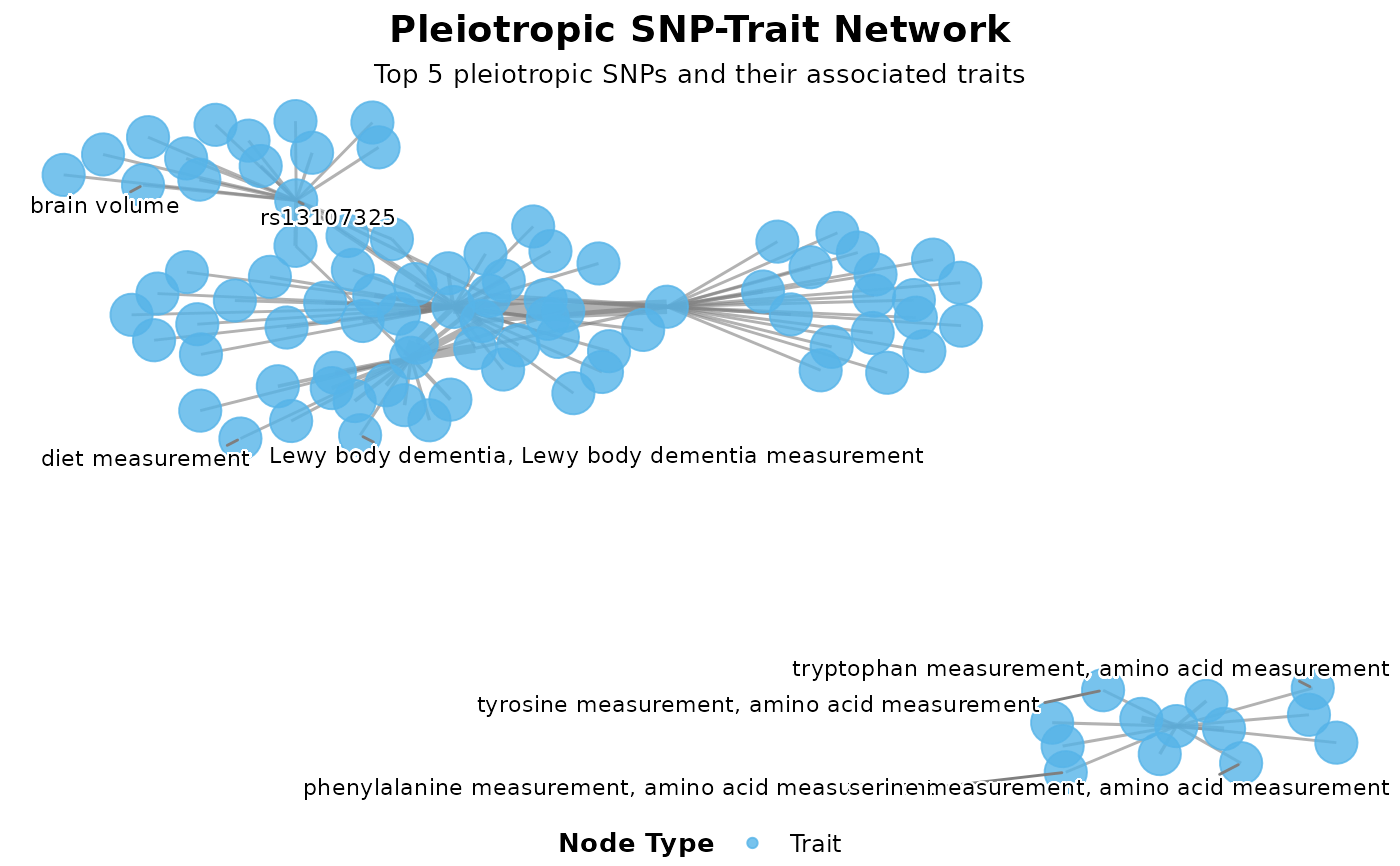
plot_pleiotropy_network.RdCreates an interactive network graph showing connections between pleiotropic SNPs and their associated traits. Nodes represent SNPs and traits, edges represent significant associations.
Usage
plot_pleiotropy_network(
pleio_data,
min_n_traits = 2,
top_n_snps = 10,
node_size_snp = 5,
node_size_trait = 8,
edge_width_max = 3,
layout = "fr",
show_labels = TRUE,
label_size = 3,
color_palette = "okabe_ito"
)Arguments
- pleio_data
A data.frame containing pleiotropy analysis results
- min_n_traits
Integer. Minimum number of traits per SNP to include (default: 2)
- top_n_snps
Integer. Number of top pleiotropic SNPs to highlight (default: 10)
- node_size_snp
Numeric. Base size for SNP nodes (default: 5)
- node_size_trait
Numeric. Base size for trait nodes (default: 8)
- edge_width_max
Numeric. Maximum edge width (default: 3)
- layout
Character. Layout algorithm: "fr", "kk", "dh", "grid", "circle" (default: "fr")
- show_labels
Logical. Show node labels (default: TRUE)
- label_size
Numeric. Label font size (default: 3)
- color_palette
Character. Color palette: "okabe_ito", "viridis", "default" (default: "okabe_ito")
Examples
data(gwas_subset)
pleio_results <- detect_pleiotropy(gwas_subset)
if (nrow(pleio_results) > 0) {
p <- plot_pleiotropy_network(pleio_results, top_n_snps = 5)
print(p)
}
#> Warning: Unknown or uninitialised column: `SNP`.
#> Warning: Unknown or uninitialised column: `SNP`.
#> Warning: ggrepel: 66 unlabeled data points (too many overlaps). Consider increasing max.overlaps