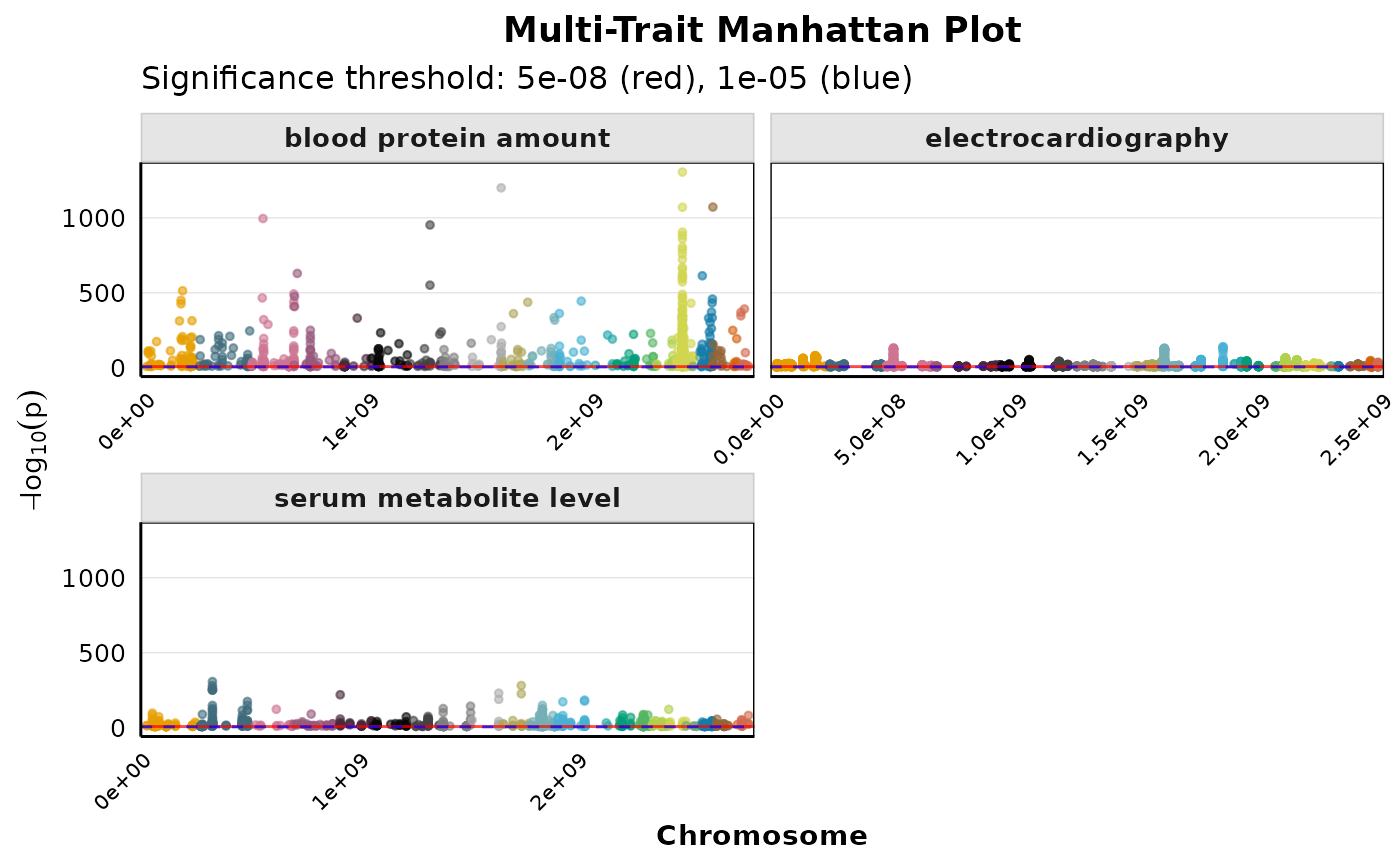
Creates a faceted Manhattan plot showing genome-wide associations for multiple traits simultaneously. Each panel represents a different trait, making it easy to compare patterns across traits.
Usage
plot_multi_trait_manhattan(
pleio_data,
traits = NULL,
max_traits = 6,
significance_line = 5e-08,
suggestive_line = 1e-05,
highlight_snp = NULL,
point_size = 1,
alpha = 0.6,
ncol = 2
)Arguments
- pleio_data
A data.frame containing pleiotropy analysis results
- traits
Character vector. Specific traits to plot (default: NULL uses top traits)
- max_traits
Integer. Maximum number of traits to display (default: 6)
- significance_line
Numeric. Genome-wide significance threshold (default: 5e-8) * @param suggestive_line Numeric. Suggestive significance threshold (default: 1e-5)
- highlight_snp
Character. SNP to highlight across all traits (default: NULL)
- point_size
Numeric. Point size (default: 1)
- alpha
Numeric. Point transparency (default: 0.6)
- ncol
Integer. Number of columns in facet grid (default: 2)
Examples
data(gwas_subset)
pleio_results <- detect_pleiotropy(gwas_subset)
if (nrow(pleio_results) > 0) {
p <- plot_multi_trait_manhattan(pleio_results, max_traits = 3)
print(p)
}